2019
Davison, Timothy; Samavati, Faramarz; Jacob, Christian
Lifebrush: painting, simulating, and visualizing dense biomolecular environments Journal Article
In: Computers & Graphics, vol. 82, pp. 232–242, 2019.
Abstract | Links | BibTeX | Tags: LINDSAY
@article{davison2019lifebrush,
title = {Lifebrush: painting, simulating, and visualizing dense biomolecular environments},
author = {Timothy Davison and Faramarz Samavati and Christian Jacob},
doi = {https://doi.org/10.1016/j.cag.2019.05.006},
year = {2019},
date = {2019-01-01},
urldate = {2019-01-01},
journal = {Computers & Graphics},
volume = {82},
pages = {232--242},
publisher = {Elsevier},
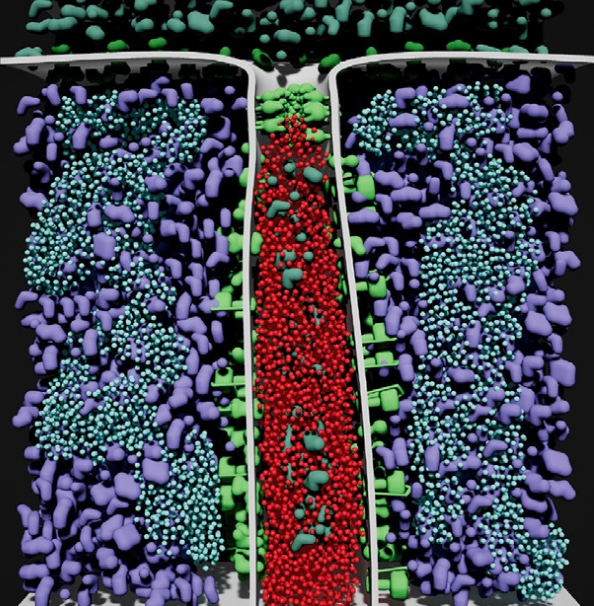
abstract = {LifeBrush is a Cyberworld for painting dynamic molecular illustrations in virtual reality (VR) that then come to life as interactive simulations. We designed our system for the biological mesoscale, a spatial scale where molecules inside cells interact to form larger structures and execute the functions of cellular life. We bring our immersive illustrations to life in VR using agent-based modelling and simulation. Our sketch-based brushes use discrete element texture synthesis to generate molecular-agents along the brush path derived from examples in a palette. In this article we add a new tool to sculpt the geometry of the environment and the molecules. We also introduce a new history based visualization that enables the user to interactively explore and distil, from the busy and chaotic mesoscale environment, the interactions between molecules that drive cellular processes. We demonstrate our system with a mitochondrion example.},
keywords = {LINDSAY},
pubstate = {published},
tppubtype = {article}
}
2018

Davison, Timothy; Samavati, Faramarz; Jacob, Christian
LifeBrush: Painting Interactive Agent-based Simulations Conference
2018 International Conference on Cyberworlds (CW), IEEE 2018.
BibTeX | Tags: game engine, LINDSAY, virtual reality
@conference{Davison2018,
title = {LifeBrush: Painting Interactive Agent-based Simulations},
author = {Timothy Davison and Faramarz Samavati and Christian Jacob},
year = {2018},
date = {2018-10-03},
urldate = {2018-10-03},
booktitle = {2018 International Conference on Cyberworlds (CW)},
organization = {IEEE},
keywords = {game engine, LINDSAY, virtual reality},
pubstate = {published},
tppubtype = {conference}
}
2013
Sarpe, Vladimir; Jacob, Christian
Simulating the decentralized processes of the human immune system in a virtual anatomy model Journal Article
In: BMC bioinformatics, vol. 14, no. 6, pp. S2, 2013.
Abstract | Links | BibTeX | Tags: immune system, LINDSAY
@article{sarpe2013simulating,
title = {Simulating the decentralized processes of the human immune system in a virtual anatomy model},
author = {Vladimir Sarpe and Christian Jacob},
doi = {10.1186/1471-2105-14-s6-s2},
year = {2013},
date = {2013-01-01},
urldate = {2013-01-01},
journal = {BMC bioinformatics},
volume = {14},
number = {6},
pages = {S2},
publisher = {BioMed Central},
abstract = {Background
Many physiological processes within the human body can be perceived and modeled as large systems of interacting particles or swarming agents. The complex processes of the human immune system prove to be challenging to capture and illustrate without proper reference to the spacial distribution of immune-related organs and systems. Our work focuses on physical aspects of immune system processes, which we implement through swarms of agents. This is our first prototype for integrating different immune processes into one comprehensive virtual physiology simulation.
Results
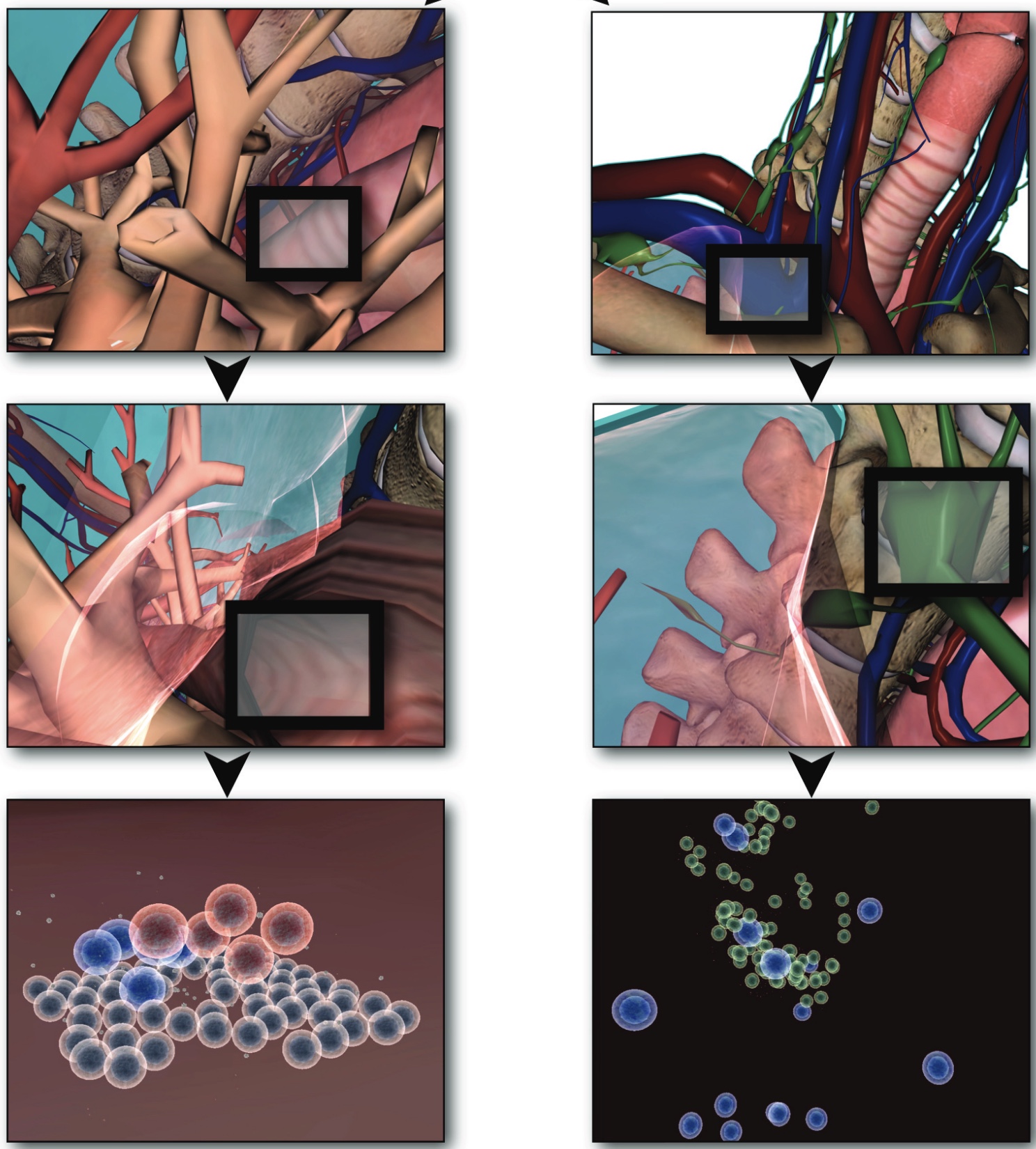
Using agent-based methodology and a 3-dimensional modeling and visualization environment (LINDSAY Composer), we present an agent-based simulation of the decentralized processes in the human immune system. The agents in our model - such as immune cells, viruses and cytokines - interact through simulated physics in two different, compartmentalized and decentralized 3-dimensional environments namely, (1) within the tissue and (2) inside a lymph node. While the two environments are separated and perform their computations asynchronously, an abstract form of communication is allowed in order to replicate the exchange, transportation and interaction of immune system agents between these sites. The distribution of simulated processes, that can communicate across multiple, local CPUs or through a network of machines, provides a starting point to build decentralized systems that replicate larger-scale processes within the human body, thus creating integrated simulations with other physiological systems, such as the circulatory, endocrine, or nervous system. Ultimately, this system integration across scales is our goal for the LINDSAY Virtual Human project.
Conclusions
Our current immune system simulations extend our previous work on agent-based simulations by introducing advanced visualizations within the context of a virtual human anatomy model. We also demonstrate how to distribute a collection of connected simulations over a network of computers. As a future endeavour, we plan to use parameter tuning techniques on our model to further enhance its biological credibility. We consider these in silico experiments and their associated modeling and optimization techniques as essential components in further enhancing our capabilities of simulating a whole-body, decentralized immune system, to be used both for medical education and research as well as for virtual studies in immunoinformatics.},
keywords = {immune system, LINDSAY},
pubstate = {published},
tppubtype = {article}
}
Many physiological processes within the human body can be perceived and modeled as large systems of interacting particles or swarming agents. The complex processes of the human immune system prove to be challenging to capture and illustrate without proper reference to the spacial distribution of immune-related organs and systems. Our work focuses on physical aspects of immune system processes, which we implement through swarms of agents. This is our first prototype for integrating different immune processes into one comprehensive virtual physiology simulation.
Results
Using agent-based methodology and a 3-dimensional modeling and visualization environment (LINDSAY Composer), we present an agent-based simulation of the decentralized processes in the human immune system. The agents in our model - such as immune cells, viruses and cytokines - interact through simulated physics in two different, compartmentalized and decentralized 3-dimensional environments namely, (1) within the tissue and (2) inside a lymph node. While the two environments are separated and perform their computations asynchronously, an abstract form of communication is allowed in order to replicate the exchange, transportation and interaction of immune system agents between these sites. The distribution of simulated processes, that can communicate across multiple, local CPUs or through a network of machines, provides a starting point to build decentralized systems that replicate larger-scale processes within the human body, thus creating integrated simulations with other physiological systems, such as the circulatory, endocrine, or nervous system. Ultimately, this system integration across scales is our goal for the LINDSAY Virtual Human project.
Conclusions
Our current immune system simulations extend our previous work on agent-based simulations by introducing advanced visualizations within the context of a virtual human anatomy model. We also demonstrate how to distribute a collection of connected simulations over a network of computers. As a future endeavour, we plan to use parameter tuning techniques on our model to further enhance its biological credibility. We consider these in silico experiments and their associated modeling and optimization techniques as essential components in further enhancing our capabilities of simulating a whole-body, decentralized immune system, to be used both for medical education and research as well as for virtual studies in immunoinformatics.
2006
Jacob, Christian; Steil, Scott; Bergmann, Karel P
The Swarming Body: Simulating the Decentralized Defenses of Immunity Conference
ICARIS, 2006.
Abstract | Links | BibTeX | Tags: biological simulation, immune system, LINDSAY
@conference{DBLP:conf/icaris/JacobSB06,
title = {The Swarming Body: Simulating the Decentralized Defenses of Immunity},
author = {Christian Jacob and Scott Steil and Karel P Bergmann},
url = {https://www.researchgate.net/profile/Christian_Jacob4/publication/221506532_The_Swarming_Body_Simulating_the_Decentralized_Defenses_of_Immunity/links/00463527b18e1c164f000000/The-Swarming-Body-Simulating-the-Decentralized-Defenses-of-Immunity.pdf},
doi = {10.1007/11823940_5},
year = {2006},
date = {2006-01-01},
urldate = {2006-01-01},
booktitle = {ICARIS},
pages = {52-65},
abstract = {We consider the human body as a well-orchestrated system of interacting swarms. Utilizing swarm intelligence techniques, we present our latest virtual simulation and experimentation environment, IMMS:VIGO::3D, to explore key aspects of the human immune system. Immune system cells and related entities (viruses, bacteria, cytokines) are represented as virtual agents inside 3-dimensional, decentralized and compartmentalized environments that represent primary and secondary lymphoid organs as well as vascular and lymphatic vessels. Specific immune system responses emerge as by-products from collective interac- tions among the involved simulated textquoteleftagentstextquoteright and their environment. We demonstrate simulation results for clonal selection and primary and secondary collective responses after viral infection, as well as the key response patterns encountered during bacterial infection. We see this simulation environment as an essential step towards a hierarchical whole-body simulation of the immune system, both for educational and research purposes.},
keywords = {biological simulation, immune system, LINDSAY},
pubstate = {published},
tppubtype = {conference}
}